Main Content
Schmeck Lab - Research
Our research focuses on:
- Lung inflammation in infection and environment-related diseases: Our research focuses on extracellular vesicles and host-pathogen interactions. We employ human cell- and tissue-models, single-cell multi-omics, along with advanced immunology and molecular biology techniques.
- Clinical and translational research: We strive to identify novel biomarkers and therapeutic strategies for lung diseases such as asthma, COPD, and pneumonia. Our clinicians are integrated into the pulmonology service of the university medical center (Universitätsklinikum Gießen und Marburg), and we support the Clinical Scientist training program at Marburg University.
- Systems biology and medicine: Our goal is to advance personalized medicine for patients with lung diseases. We analyze extensive clinical and experimental data, as well as the complexities of human diseases, utilizing high-throughput screening, bioinformatics, and mathematical modeling.
Ongoing projects on infectious diseases and host-pathogen interaction:

|
LOEWE Diffusible Signals (funded by the Hessian Ministry of Research/HMWK) Collaborative research center on diffusible signals in host-pathogen interaction. Universities of Marburg and Giessen and Max-Planck Institute for Terrestrial Microbiology |

|
LOEWE Exploration (funded by the Hessian Ministry of Research/HMWK) Research project on antimicrobial peptides in infectious diseases. Universities of Marburg and Muenster. |
|
Deep Legion (funded by the German Ministry of Research/BMBF) Research projects on Legionella pneumophila pathobiology. Universities of Marburg and Muenster |

|
German Center for Lung Research/DZL (funded by the German Ministry of Research/BMBF) |
|
PROGRESS (funded by the German Ministry for Health/BMG) Clinical Research Center on longCOVID. University of Marburg. |
|
|
|
Ongoing projects on chronic and environment-related lung diseases:

|
Permed-COPD (funded by the German Ministry of Research/HMWK) German Research Platform to implement Personalised Medicine of COPD. Universities of Marburg, Muenster, Heidelberg, and Munich. |

|
LOEWE HABITAT (funded by the Hessian Ministry of Research/HMWK) Collaborative Research Center on the impact of weather and climate on human health. Universities of Marburg and Fulda |

|
CALM-QE (funded by the German Ministry of Research/BMBF) Research Center for Medical Informatics with focus on COPD and Asthma. |

|
German Center for Lung Research/DZL (funded by the German Ministry of Research/BMBF) |

|
3TR (funded by the European Union) European Research Network on Biological Treatments of chronic inflammatory diseases |
|
|
Funded Research Projects
LOEWE Diffusible Signals

How do bacteria communicate with human inflammatory cells? This topic is at the center of the LOEWE research cluster "Diffusible Signals" (Impact of diffusible signals at human cell-microbe interfaces). The Hessian state government is funding "Diffusible Signals" in the 13th season of the Hessian state offensive for the development of scientific and economic excellence ("Landesoffensive zur Entwicklung wissenschaftlicher und ökonomischer Exzellenz" - LOEWE) with a total of about 4.8 million euros.
Subproject A2 - Diffusible Signals as Colonization Factors

Cholera is a devastating intestinal disease caused by the bacterium Vibrio cholerae. Contrary to a long-held belief, V. cholerae has recently been shown to trigger an inflammatory response during infection. However, it is unclear how V. cholerae cells interact with the innate immune system. In preliminary work, we have investigated the interaction with macrophages and found that the diffusible protein TcpF secreted by V. cholerae is essential for the binding of V. cholerae to macrophages and may have an influence on the secretion of diffusible cytokines. Here, we want to elucidate the mechanism by which TcpF can cause or facilitate the attachment of V. cholerae to macrophages and how macrophages react to TcpF. Understanding the detailed mechanism of this important step in the interaction between V. cholerae and macrophages could lead to new therapeutic approaches against cholera.
Principal Investigators:
Knut Drescher
Bernd Schmeck
Subproject B1 - Extracellular Vesicles in Host-Microbe-Interactions

Recent results indicate that both the extracellular vesicles (EVs) of host cells and the outer membrane vesicles (OMVs) of microbes may have an influence on the course of inflammation and infection. Our own preliminary work has shown cell-specific inflammatory effects of EVs. In addition, replication-promoting as well as replication-inhibiting properties of Legionella pneumophila OMVs have been observed.
The aim of project B1 is to comprehensively characterize the functional components and effects of both diffusible vesicle types using the model of Klebsiella (Kp)-macrophage (MΦ) interaction. By analyzing the origin and composition of EVs in the infection context as well as the response of host cells and bacteria thereto (recognition, uptake, utilization, signal transduction), we will significantly broaden our understanding of cell and infection biology. At the same time, the project will also develop the basis for a clinical-translational use of vesicles in diagnostics, prevention, and therapy of bacterial infectious diseases.
Principal Investigators:
Bernd Schmeck
Anna Lena Jung
The projects are funded by the Hessisches Ministerium für Wissenschaft und Forschung – LOEWE.
LOEWE Exploration - Analysis of Human Antimicrobial Resistance Memory with Artificial Intelligence as a Strategy against Microbial Resistance

According to the WHO, the increase in antimicrobial resistance (AMR) is one of the greatest threats to global health, food security, and social development. It is estimated that the number of annual deaths worldwide will reach 10 million by 2050 if no action is taken to combat AMR. New active ingredients are urgently needed. Antimicrobial peptides (AMPs) are part of the inherent immune system of almost all organisms and represent a potent alternative to conventional antibiotics as they are less likely to lead to the development of resistance. Even though the first AMPs are already being used clinically, there is a lack of detailed knowledge about the regulation of the specific AMP response during infection. Among other things, it is not clear whether AMP-secreting cells are capable (similar to other cells of the innate immune response) of forming a short-term memory (trained immunity) in order to release pathogen-specific AMPs more quickly after reinfection. Our aim is to use in vitro infection experiments and bioinformatic analyses to analyze the time course of AMP expression in order to explore whether cells can develop this trained immunity.
Our project is divided into three phases: (i) performing infection series in human cells and generating transcriptional and mass spectrometry profiles, (ii) bioinformatic analysis of the specific and time-dependent AMP response using innovative AI-based approaches, and (iii) in vitro validation. If we find evidence for a cellular AMP memory, this result may be an important starting point for targeted therapy and drug development.
Principal Investigators:
Bernd Schmeck
Dominik Heider
Funded by the the Hessisches Ministerium für Wissenschaft und Forschung, LOEWE Exploration.
Deep Legion Detection of virulence factor protein domains in Legionella using deep autoencoders

Many of these factors are essential virulence factors and, therefore, important for understanding disease pathophysiology and potential therapeutic targets. Thus, to improve the treatment for patients, the accurate annotation of these sequences is of utmost importance.
This project aims to identify these virulence factors computationally based on our current data, computationally predict their origin and function, and validate these predictions in vitro. Since there are many limitations with existing tools, we aim to develop a deep learning-based framework that can be used to identify such virulence factors in almost real-time with high accuracy. Our deep learning-based annotation pipeline will pave the way to new applications for precision medicine in infectious diseases.
We will implement this new deep learning-based annotation pipeline as an ultrafast bioinformatics command-line software tool throughout this project. Furthermore, we will complement this tool by database creation features for the compilation of customized databases. By doing so, we will provide the bioinformatics community with a powerful tool for the rapid annotation of DNA sequences, protein motifs, and domains. Additionally, we will containerize and provide this tool within highly scalable cloud computing infrastructures.
Principal Investigators:
- Dominik Heider
- Alexander Goesmann
- Bernd Schmeck
Funded within the BMBF initiative on Computational Life Sciences 2021.
PerMed-COPD

The overall goal of PerMed-COPD is to improve COPD diagnosis and classification in clinical practice in a new personalized medicine platform. For that, we perform a prospective trial to translate recent research results and previous joint work on imaging and molecular biomarkers as well as clinical decision support systems (CDSS) and artificial intelligence.
The main objectives are:
With the help of a clinical study and an economic evaluation, we validate a previously developed therapeutic decision aid based on objectified diagnostic assessment markers, i. e., imaging biomarkers, focusing on comorbidities of COPD. In addition, we use our prospective diagnostic study for the prognosis of the disease course and for the prediction of therapy response.
Using deep genotyping and phenotyping, we define clinically relevant subgroups and develop innovative integrated analysis tools, which enable new perspectives on molecular disease mechanisms, development and progression of diseases, as well as therapy.
Based on the interpretation of multi-factorial and multi-modal data, we implement and optimize an innovative and user-friendly clinical decision support for medical professionals.
We enable personalized treatment strategies using comprehensive integration of various data sources such as clinical data, medical imaging, and omics data as well as machine learning.
Subproject 3: Molecular Biomarkers
COPD represents a major medical challenge due to its great heterogeneity with multiple comorbid traits and diseases. Therefore, we carry out biologically-driven patient stratification based on polygenic risk scores to clinically define the relevance of different biomarkers (whole blood transcriptomics, extracellular vesicle surface proteomics, sputum microbiome) and to validate their correlation with the progression of the disease.
In our research project, we make use of existing clinical data to detect clinically relevant biomarkers and to validate them in a prospective, multi-center validation cohort. We take into account both technological feasibility as well as regulatory and commercial documentation and quality management standards to speed up technology transfer into translation and regulatory approval. The expertise we gain from these experiments aids in pre-preparatory work for the regulatory approval and clinical application that represent a major step towards a personalized medicine for COPD prevention and treatment.
Principal Investigators:
- Bernd Schmeck
- Dominik Heider
- Johannes Schumacher
The project is funded by the BMBF.
HABITAT – Health Affected by Climate Change and Air Pollution

Climate change has been recognized by the World Health Organization (WHO) as the greatest threat to human health. At present, however, there is no comprehensive knowledge of weather- and environment-related acute illnesses, no personalized warning systems for people affected by extreme weather events and no corresponding organization of the healthcare system. Extreme weather events lead to increased health risks, especially for people who are already exposed, and thus to unpredictable burdens on medical care in the short term. The aim of the research network HABITAT is to develop a functioning forecasting system for predicting individual risks in interdisciplinary cooperation. We involve artificial intelligence in order to reduce the risks of acute episodes of illness through behavioral adjustments and to be able to predict and plan the allocation of resources in the healthcare system.
Subproject A: Influence of Weather and Environmental Factors on Disease Progression
In subproject A, we study the effects of weather events and environmental factors in interaction with personal characteristics on the incidence, prevalence, and pathogenesis of diseases. As use cases, we focus on heart failure, including acute cardiac decompensation to cardiogenic shock, coronary heart disease, as well as tachycardic cardiac arrhythmias, chronic obstructive pulmonary disease (COPD), bronchial asthma, pneumonia, and hypertensive diseases during pregnancy. In order to investigate the effects of environmental and weather events on these diseases, existing patient databases are linked with regional weather and environmental data from the region of Marburg and Fulda (Mittelhessen). We can rely on the expertise, technical equipment (e. g., own operational weather stations with high temporal resolution), and the well-established cooperation of our universities. In addition, we use openly accessible data from other organizations, e. g., the Deutscher Wetterdienst or the Hessisches Landesamt für Naturschutz, Umwelt und Geologie (HLNUG) to incorporate weather data, air quality data, and pollen data into the models.
Principal Investigators:
Jörg Bendix
Thomas Brenner
Mareike Lehmann
Corinna Keil
Bernd Schmeck
Bernhard Schieffer
The project is funded by the Hessisches Ministerium für Wissenschaft und Forschung – LOEWE.
CALM-QE - Multidimensional Asthma & COPD Research Models Based on AI and Machine Learning

Innovative IT technology is intended to help doctors diagnose chronic respiratory diseases, such as ASTHMA and COPD, more precisely and treat each individual case in a tailor-made manner. This approach is sometimes complicated because the mechanisms of these diseases vary from case to case. The result: What provides relief for one person does not necessarily help another.
In CALM-QE, we therefore use specially developed computer programs to analyze data from health care and to learn how the different variants of chronic respiratory diseases can be assigned to individual patients more precisely – because the more precisely the individual diagnoses can be made, the more targeted and efficient they are those affected can be treated.
Subproject 3: Data from Outpatients in Private Pracice and Cross-Sector Longitudinal Follow-up
Chronic obstructive pulmonary disease (COPD) and bronchial asthma (BA) are the most common non-communicable pulmonary diseases with a major socio-economic impact. Development, severity and exacerbations are the result of complex gene-environment interactions. Recent research revealed a broad heterogeneity in terms of phenotypes and endotypes with some degree of overlap between COPD and BA. This novel concept is paralleled by the development of more specific medications, leading to the challenge to stratify patients for this precision medicine approach. The major challenge in the field of COPD and BA is how to translate this treatable trait approach to the individual patient. Therefore, it is the goal of CALM-QE to develop, train and test predictive models for important clinical outcomes based on multidimensional “real-world-datasets” for COPD and BA patients across sectoral boundaries. They include the already established clinical core dataset, expansion by lung function, medication, chest imaging, addition of local climate and air pollutant data, and biosignals obtained via wearables. All of these data will be used to identify and group disease trajectories. In addition, as an initial proof of concept, we will incorporate research study data, including longitudinal data from the German COPD cohort. As both COPD and BA originate in early life, we will model life-spanning trajectories from childhood into adulthood. Furthermore, most of the patients are regularly seen in private practices. Thus, the project incorporates cross-sectorial health care providers ranging from private practices to university hospitals. In order to accomplish this goal, an interdisciplinary team of experts from pediatric and adult medicine, epidemiology, and medical informatics has been formed across 12 participating university hospitals and partnering practices, based on the strong foundation of local infrastructures such as data integration centers. In conclusion, the novel predictive models built by the project from multi-dimensional large datasets for clinically relevant endpoints will serve the 4P medicine approach (personalized, participatory, predictive, and preventive).
Principal Investigators:
Christian Taube
Harald Renz
Bernd Schmeck
Anna-Maria Dittrich
This project is funded by the Medizin Informatik Initiative des BMBF.
More Projects
Potential of Extracellular Vesicles as Biomarkers, Trigger, and Therapeutic Aid in COPD

Chronic obstructive pulmonary disease (COPD) is caused by longterm exposure to inhaled substances like cigarette smoke and currently forms the third leading cause of death worldwide. COPD management is complicated by the fact that COPD patients are highly heterogeneous in terms of pathophysiology, changes to the lung tissue, and comorbidities. For this reason, the current clinical criteria do not allow predictive patient stratification and no disease-modifying therapeutics are currently available. In order to address the strong need to identify biomarkers for patient stratification, elucidate pathomechanisms underlying disease development and progression in (subgroups of) COPD patients, and establish novel therapeutic strategies, our project aims at establishing a quick test for COPD patient stratification on circulating EVs, determining the effect of EVs on pathophysiology, and validating engineered EVs for targeted intracellular mRNA delivery.
Principal investigators:
- Bernd Schmeck
- Birke Benedikter
- Nurlan Dauletbayev
The project is funded by the University of Marburg, University of Giessen and CSL Research Acceleration Initiative .
Sepsis-Induced Makrophage-Endothel-Interaction: Bacterial Extracellular Vesicles as Regulators of RNase1
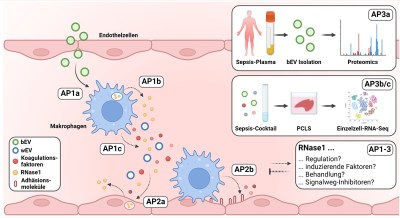
Sepsis is the most common cause of infection-related death worldwide and tightly associated with a dysregulation of the immune response, a coagulation-promoting activation of the vasculature and associated endothelial dysfunction. In this context, ribonuclease 1 (RNase1) acts as an important protective factor of vascular homeostasis, whose function is impaired after massive and persistent inflammation which promotes the development of vascular diseases. Bacterial extracellular vesicles (bEVs) are secreted by pathogens in the context of infection and current studies in our group showed that bEVs are significantly involved in the repression of endothelial cell-derived RNase1. In this project, we further investigate the influence of bEVs on RNase1 in the context of sepsis in more detail. The effect of bEVs on the regulation of RNase1 and coagulation-promoting factors in human macrophages as well as the influence of these factors on the inflammatory response and interaction with the lung endothelium will be investigated. Furthermore, the isolation of bEVs from human plasma will be established to analyze the proteome of bEVs in the plasma of sepsis patients. In addition, these findings will be used to establish a human ex vivo-sepsis tissue model to further define the role of RNase1 in sepsis-induced tissue damage using single-cell sequencing and molecular biological analyses.
Principal Investigator:
- Katrin Laakmann
The project is funded by the Universitätsklinikum Gießen und Marburg – UKGM.
Modulation of T Helper Cell Immunity by Extracellular Vesicles in Bacterial Pneumonia
Pneumoniae are a continual challenge in clinical practice. The persistent occurrence of multidrug-resistant Pseudomonas aeruginosa contributes to a high motility rate, especially in pneumonia and bloodstream infections, and in burn patients. In this project, we investigate the immunomodulatory properties of extracellular vesicles (EVs) produced by monocytes/macrophages and dendritic cells during P. aeruginosa infection. T cells particularly T helper cells, respond in dispensable bacterial clearance and host survival. In light of preliminary data, we specifically investigate the effect and target of pro-inflammatory cytokine and of the major histocompatibility complex (MHC) transporting EVs in the context of adaptive immune responses. The study addresses whether these early infection-derived immune cell EVs affect helper T cell activation and differentiation, thereby initiating a strong immune response. The projects aims at understanding the early infection process from a previously unexplored perspective. To investigate the physiological relevance, the study will also detect such immunomodulatory EVs in an in vivo P. aeruginosa-mice-infection model and in patient samples. Deciphering the underlying processes will aid in the prediction and development of antibiotic-independent therapeutics.
Principal Investigators:
- Bernd Schmeck
- Ulrich Matt
Funded by the Universitätsklinikum Gießen und Marburg – UKGM.
Extracellular Vesicles as Clinical and Pathophysiological Biomarkers in Acute Respiratory Infections
Acute respiratory infections are the fourth leading cause of death worldwide. Having characterized the pathophysiological influences of extracellular vesicles (EVs) in experimental infection studies and the molecular changes in clinical pilot studies on malaria and other infectious diseases, we postulate that EVs in patient plasma are suitable biomarkers for disease progression of acute lower respiratory tract infections and the underlying pathophysiology. In a two-year pilot study, we will recruit patients with acute lower respiratory tract infections and isolate and characterize EVs from their plasma.
Principal Investigators:
- Philipp Markart
- Bernd Schmeck
This project is funded by the Ausschreibung Forschungsföderung Campus Fulda.