Main Content
Biosynthesis of cellular iron-sulfur proteins and the impact on cellular iron homeostasis
Our cell biological and biochemical research concentrates on the molecular mechanisms underlying the biogenesis of iron-sulfur (Fe/S) proteins in eukaryotes, a process we discovered in 1999. Initial studies were performed in baker’s yeast Saccharomyces cerevisiae, but later investigations showed the strong conservation of the pathway from yeast to human cells. Fe/S proteins are located in mitochondria, cytosol and nucleus, where they participate in electron transfer reactions, enzyme catalysis, sulfur transfer, and regulatory reactions (Figure 1). Numerous important cellular processes depend on Fe/S protein function, including mitochondrial respiration, citric acid cycle, metabolic reactions, DNA synthesis and repair, ribosome biogenesis, tRNA modification, viral defense, and iron uptake regulation. Hence, Fe/S clusters are not only ancient protein cofactors thought to have played a decisive role in the origin of life, but are also indispensable for most living organisms.
Figure 1. Simplified overview of the localization, function and biogenesis of cellular iron-sulfur proteins.
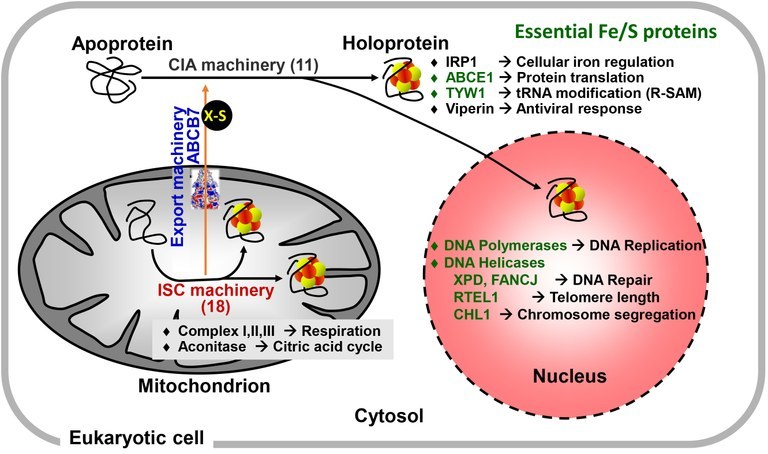
Known iron-sulfur (Fe/S) proteins are localized in mitochondria, cytosol and nucleus. They are involved in numerous cellular functions and pathways. Some (shown in green) execute essential functions by participating in, e.g., protein translation or DNA replication and repair. Classical Fe/S proteins are the mitochondrial aconitase and respiratory complexes I, II, and III, whereas viperin involved in antiviral response has been discovered more recently. The synthesis and insertion of Fe/S clusters in living cells needs complex machinery which is highly conserved in eukaryotes from yeast to man. The mitochondria-localized iron-sulfur cluster assembly (ISC) machinery comprises 18 ISC proteins, and is involved in the biogenesis of all cellular Fe/S proteins. The core ISC proteins assist the cytosolic iron-sulfur protein assembly (CIA) machinery (11 known proteins) by synthesizing an unknown sulfur-containing molecule (X-S) which is exported to the cytosol by the ABC transporter ABCB7 (Atm1 in yeast). Mitochondrial Fe/S protein biogenesis is tightly linked to cellular iron regulation by its role in the maturation of iron regulatory protein 1 (IRP1) in humans.
The assembly of [2Fe-2S] and [4Fe-4S] clusters and their insertion into apoproteins in a living cell is a complex process catalyzed by more than 30 known biogenesis proteins in mitochondria and cytosol (Figure 1). In mitochondria, the iron-sulfur cluster assembly (ISC) machinery was inherited from bacteria by endosymbiosis. The ISC system is involved in biogenesis of all cellular Fe/S proteins. The core part of the ISC machinery also generates a sulfur-containing factor (X-S in Figures) which is exported to the cytosol via the mitochondrial ABC transporter ABCB7. There, X-S is used by the cytosolic iron-sulfur protein assembly (CIA) machinery composed of 11 known proteins to generate cytosolic and nuclear Fe/S proteins. Mechanistic details of these processes are explained in Figure 2.
Knowledge of molecular aspects of Fe/S protein biogenesis is of medical interest. Genetic mutations in virtually all ISC components are causative of so-called ‘Fe/S diseases’ which are characterized by diverse metabolic, neurological and hematological phenotypes. The founding members of these rare diseases are the iron storage disorders Friedreich’s ataxia (FRDA) and X-chromosome-linked sideroblastic anemia and cerebellar ataxia (XLSA/A) caused by mutations in human frataxin and ABCB7, respectively (see below, Figure 2). Mutations in the late-acting mitochondrial ISC components (green in Fig. 2) cause the ‘Multiple Mitochondrial Dysfunction Syndromes’ (MMDS). Moreover, functional defects in Fe/S proteins involved in DNA repair (e.g., XPD and FANCJ DNA helicases; Figure 1) are connected to multiple diseases such as skin cancer and Fanconi anemia.
Most components of eukaryotic Fe/S protein biogenesis are essential for cell viability. Unexpectedly, the indispensable character of the mitochondrial components is explained by their crucial function in the maturation of extra-mitochondrial rather than mitochondrial Fe/S proteins. One of the essential cytosolic Fe/S proteins (human ABCE1) performs a central role in protein translation, namely the splitting of the ribosomal subunits after translation termination (Figure 1). Other translation-related cytosolic Fe/S proteins (TYW1, ELP3) are responsible for various base modifications in tRNAs. In the nucleus, DNA polymerases and ATP-dependent DNA helicases involved in DNA synthesis and repair depend on Fe/S clusters for function. The involvement of mitochondria in the generation of these essential cytosolic and nuclear Fe/S proteins (and not mitochondrial ATP synthesis by oxidative phosphorylation) explains the organelle’s indispensable character within the eukaryotic cell. Moreover, biogenesis of cellular Fe/S proteins by the ISC machinery appears to represent the minimal function of mitochondria. This is biologically supported by the discovery of ‘mitosomes’ in, e.g., Microsporidia, Giardia, and Cryptosporidium. During evolution these simplified mitochondria-related organelles (MRO) have lost almost all classic mitochondrial functions, yet they maintained the core ISC system as their residual biochemical function.
Figure 2. Detailed working model for the maturation of cellular iron-sulfur proteins in human cells.
![The biogenesis of iron-sulfur (Fe/S) proteins is conserved from yeast to man, and in mitochondria requires the ISC machinery with 18 known proteins. The mitochondrial ISC pathway can be dissected into three main steps. First, a [2Fe-2S] cluster is de novo assembled on the scaffold protein ISCU. Synthesis of the cluster requires the cysteine desulfurase complex NFS1-ISD11-ACP1 releasing sulfur from cysteine and transferring it to ISCU. Frataxin is proposed to support sulfur transfer and/or supply iron, while the sulfur reduction to sulfide requires the electron transfer chain NADPH-ferredoxin reductase-ferredoxin (FDXR, FDX2). Reduced iron (Fe2+) is imported into mitochondria by the carrier proteins mitoferrin1/2 in a proton-motive force (pmf) dependent fashion. In the second step, the ISCU-bound [2Fe-2S] cluster is released from ISCU by direct binding of dedicated Hsp40- and Hsp70 chaperones (HSC20, HSPA9). The [2Fe-2S] cluster is transferred to glutaredoxin GLRX5, from where it is directly inserted into [2Fe-2S] apoproteins completing their maturation. In the final step, the synthesis of [4Fe-4S] clusters is catalyzed by fusing the GLRX5-bound [2Fe-2S] clusters, a reaction involving the specialized ISC proteins ISCA1-ISCA2-IBA57. While insertion into apoproteins such as aconitase does not need further assistance, complex Fe/S proteins such as respiratory complexes I and II and the radical SAM protein lipoyl synthase (LIAS) require further, late-acting ISC factors for Fe/S cluster transfer. For instance, IND1 (also termed NUBPL) or NFU1 both transiently bind the [4Fe-4S] cluster during its trafficking to recipient apoproteins.](https://www.uni-marburg.de/de/fb20/bereiche/zyto/bilder/bilder-lill/image_large-1.jpg/@@images/image/large)
The biogenesis of iron-sulfur (Fe/S) proteins is conserved from yeast to man, and in mitochondria requires the ISC machinery with 18 known proteins. The mitochondrial ISC pathway can be dissected into three main steps. First, a [2Fe-2S] cluster is de novo assembled on the scaffold protein ISCU. Synthesis of the cluster requires the cysteine desulfurase complex NFS1-ISD11-ACP1 releasing sulfur from cysteine and transferring it to ISCU. Frataxin is proposed to support sulfur transfer and/or supply iron, while the sulfur reduction to sulfide requires the electron transfer chain NADPH-ferredoxin reductase-ferredoxin (FDXR, FDX2). Reduced iron (Fe2+) is imported into mitochondria by the carrier proteins mitoferrin1/2 in a proton-motive force (pmf) dependent fashion. In the second step, the ISCU-bound [2Fe-2S] cluster is released from ISCU by direct binding of dedicated Hsp40- and Hsp70 chaperones (HSC20, HSPA9). The [2Fe-2S] cluster is transferred to glutaredoxin GLRX5, from where it is directly inserted into [2Fe-2S] apoproteins completing their maturation. In the final step, the synthesis of [4Fe-4S] clusters is catalyzed by fusing the GLRX5-bound [2Fe-2S] clusters, a reaction involving the specialized ISC proteins ISCA1-ISCA2-IBA57. While insertion into apoproteins such as aconitase does not need further assistance, complex Fe/S proteins such as respiratory complexes I and II and the radical SAM protein lipoyl synthase (LIAS) require further, late-acting ISC factors for Fe/S cluster transfer. For instance, IND1 (also termed NUBPL) or NFU1 both transiently bind the [4Fe-4S] cluster during its trafficking to recipient apoproteins.
The maturation of cytosolic and nuclear Fe/S proteins depends on the core ISC system providing sulfur from mitochondrial NFS1 and generating the unknown sulfur-containing compound X-S. The mitochondrial ABC transporter ABCB7 exports X-S to the cytosol in a glutathione (GSH)-dependent step, where it is used by the CIA machinery for the synthesis of a [4Fe-4S] cluster on CFD1-NBP35 P-loop ATPases serving as a scaffold complex. The latter reaction needs electron input from the flavin-oxidoreductase NDOR1 and the Fe/S protein CIAPIN1 (yeast Tah18 and Dre2). The complex of GLRX3-BOLA2 facilitates Fe/S cluster insertion into CIAPIN1. The CFD1-NBP35-bound [4Fe-4S] cluster is then released and trafficked to the majority of cytosolic and nuclear target Fe/S apoproteins by the help of CIAO3 (yeast Nar1), and the CIA targeting complex (CIAO1, CIAO2B, and MMS19) which facilitates cluster insertion by direct apoprotein interaction. Interestingly, the translation-associated ATPase ABCE1 additionally needs the ORAOV1‑YAE1D1 adapter complex (yeast Lto1‑Yae1) to recruit the apo-ABCE1 to the CIA targeting complex for [4Fe-4S] cluster transfer. Maturation of the iron regulatory protein 1 (IRP1) is unique in that it depends on the early part of the CIA machinery, yet - instead of the CIA targeting complex – it uses CIA2A (a close homolog of CIA2B) for Fe/S cluster insertion, thereby linking cellular Fe/S protein biogenesis and cellular iron regulation.
Since Fe/S protein biogenesis is tightly connected to cellular and mitochondrial iron uptake and regulation, our group also studies these processes. Defects in the core mitochondrial ISC components or ABCB7 result in increased cellular iron uptake and iron accumulation in mitochondria. To better understand the molecular basis underlying the (dys)regulation of cellular iron compartmentation as a result of Fe/S protein defects, we examine the components and mechanisms involved in mitochondrial iron transport and regulation. We previously found that mitochondrial iron import requires reduced (ferrous) iron, a membrane potential, and the yeast mitochondrial carrier proteins Mrs3-Mrs4 (human mitoferrin1/2; Figure 2).
We are grateful for generous financial support by