Main Content
Introduction
Every cell in our body contains the genetic information to produce the proteins for specific biological tasks. To assure that a cell only produces the proteins that are necessary for its cellular function, the expression of each gene is tightly regulated. Transcription is the first and most critical step for gene expression. The transcription level of a gene is determined by a complex interplay of DNA binding transcription factors and the RNA Polymerase II. The process of gene regulation also involves the so-called chromatin, whose packaging influences the transcriptional process. The property of the chromatin at specific loci is modulated by chromatin regulatory proteins, such as histone modifying enzymes and chromatin remodellers.
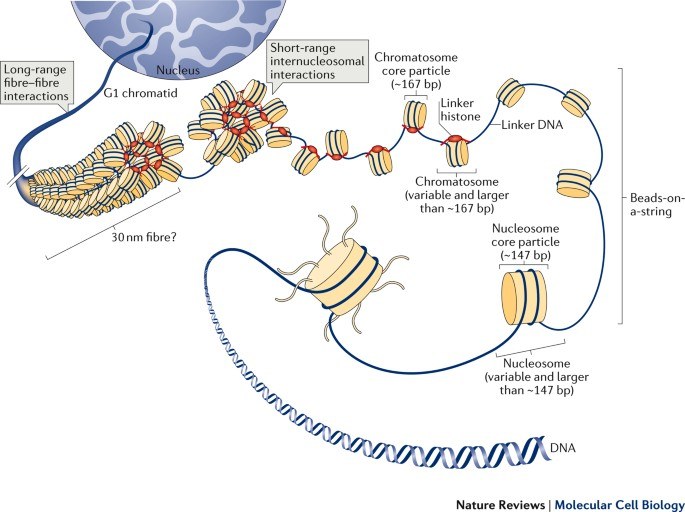
Fyodoro et al. 2018, Nature Reviews Molecular Cell Biology
The main interest of our lab is to understand molecular processes that are involved to regulate the chromatin status and transcription, with a particular focus on proteins and protein complexes that inhibit transcriptional activity. Further, we investigate the role of those proteins in cancer and we aim to develop novel strategies to modulate their functions in cancer.
Projects
Molecular mechanisms of SAMD1 and its role in human cancer
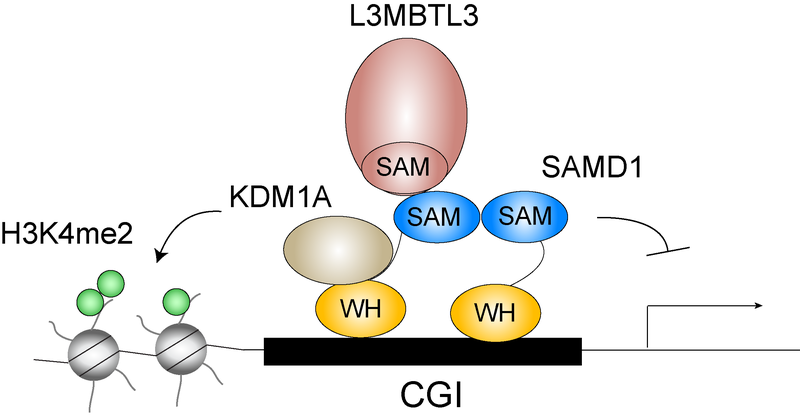
Our lab identified and characterized the protein SAMD1 (SAM domain containing protein 1) as a protein that specifically binds to so called CpG islands (CGIs) in the genome. SAMD1 possesses a DNA binding winged helix (WH) domain and a SAM domain, which allows polymerization. Both the DNA binding and the polyerimerization ability is important for the chromatin binding of SAMD1. At its target genes SAMD1 preferentially acts as a transcriptional repressor via interacting with repressive proteins including the histone demethylase KDM1A and the chromatin binding protein L3MBTL3. SAMD1 is essential for proper gene expression, and its absence leads to embryonic lethality.
Currently, our major interest is to understand how SAMD1 function is precisely regulated, and how SAMD1 in turn is involved to regulate the transcription of its target genes. SAMD1 is also commonly dysregulated in various cancer types, and its expression often correlates with patient survival. Using human cancer cell lines as model it is our goal to elucidate the role of SAMD1 in specific cancer types, such as pancreatic cancer and AML.
This project is performed in collaboration with Zhanxin Wang (Beijing University, China), Magdalena Huber, Thorsten Stiewe, Andreas Neubauer and Matthias Lauth (University of Marburg)
References:
Campbell and Weber et al., 2023, Scientific Reports (PMID: 36810619)
Simon et al., 2022, Biology (Basel), (PMID: 35453756)
Stielow and Zhou et al., 2021, Science Advances, (PMID: 33980486)
Stielow et al., 2021, Comput Struct Biotechnol J, (PMID: 34136100)
Understanding how dysregulation of KAT6A/B contributes to developmental syndromes and AML
Recently, we also identified novel WH domains in the histone acetyltransferase KAT6A and KAT6B, which have a similar DNA binding function as the WH domain in SAMD1. Consequently, we could demonstrate that KAT6A is also recruited to unmethylated CpG islands. This project was performed in cooperation with Zhanxin Wang (Beijing, China) and Martha Bulyk (Boston, USA).
Importantly, mutations/rearrangements that involve KAT6A/B are known to lead to developmental syndromes and to acute myeloid leukemia (AML). In most of these aberrations the DNA binding domain remains intact, suggesting that the mutated proteins are recruited to similar places as the wildtype proteins. In our future work we aim to better understand how the modified KAT6A/B proteins impact gene transcription, and how this may contribute to human diseases.
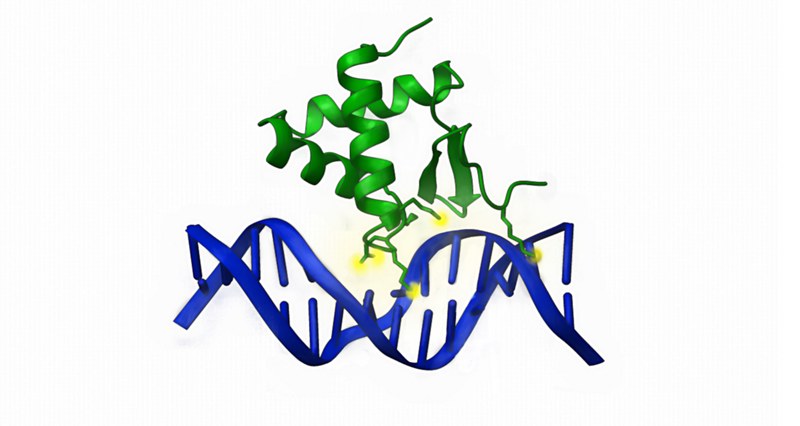
Reference:
Weber and Jia et al., 2023, Nucleic Acids Research (PMID: 36537216)
Molecular mechanisms of IRF8 and the IRF2BP2-complex and their roles in acute myeloid leukemia
We and others have identified the inflammation related proteins IRF8 (interferon regulatory factor 8) and IRF2BP2 (IRF2 binding protein 2) as particularly important in acute myeloid leukemia (AML). IRF8 and is a DNA binding transcription factor, while IRF2BP2 forms together with IRF2BP1 and IRF2BPL a trancriptional corepressor complex.
We are currently particularly interested to understand how the IRF2BP2-complex is recruited to chromatin in AML cells, and by which molecular processes the IRF2BP2-complex facilitates gene repression.
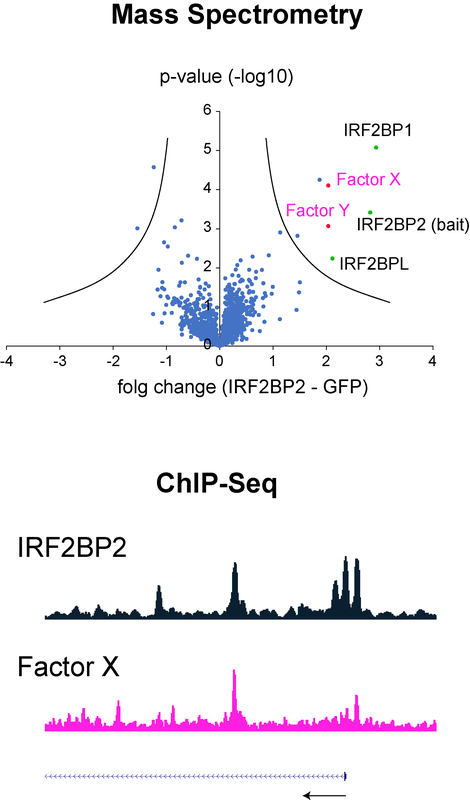
This project is performed in collaboration with Andreas Neubauer, Miriam Frech and Andreas Burchert (University of Marburg)
Reference:
Fischer et al. 2023, Preprint
Liss et al., 2021, Cancers (Biology) (PMID: 33673123)
Development of an inhibitor for Elongin BC
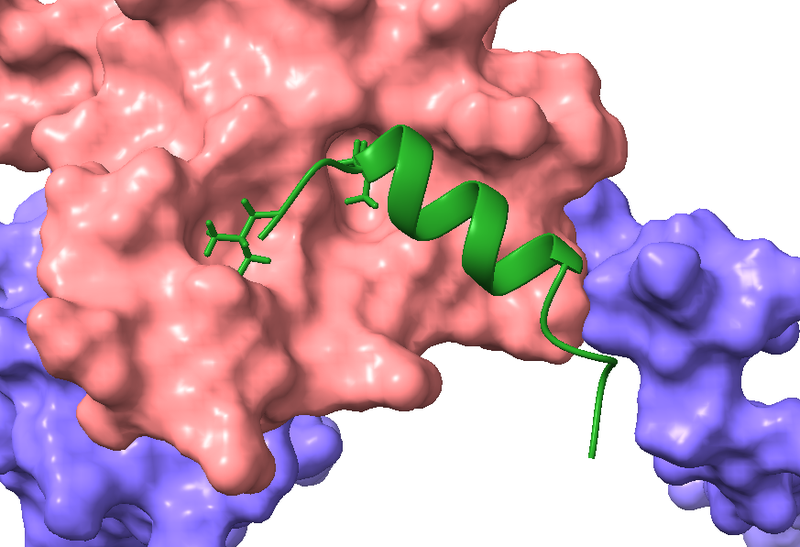
The heterodimer Elongin BC is involved in transcriptional regulation and protein turnover. It is essential for cancer cell growth, making this dimer an attractive drug target. Together with the lab of Olalla Vázquez (Department of Chemistry, University of Marburg) we are developing inhibitors for Elongin BC. Previous work has shown that Elongin BC interacts with the Polycomb repressive complex 2-associated protein EPOP via a specific sequence, called the BC-box. Using the BC-box of EPOP as starting point, we created peptide inhibitors for Elongin BC. Using this peptide, we confirmed that Elongin BC is inhibitable in cells, demonstrating the feasability of this approach. In the future we aim to further optimize the Elongin BC inhibitors to perturb the function of Elongin BC in cancer and to gain new insights into the cellular role of Elongin BC.
References:
Fischer and Trinh et al, 2022, Cell Chemical Biology (PMID: 37354906)
Liefke et al., 2016, Molecular Cell (PMID: 27863226)