Main Content
Area D - RNA Mechanisms
D1 - Influence of Diffusible Metabolites on Infection
PI: Lennart Randau
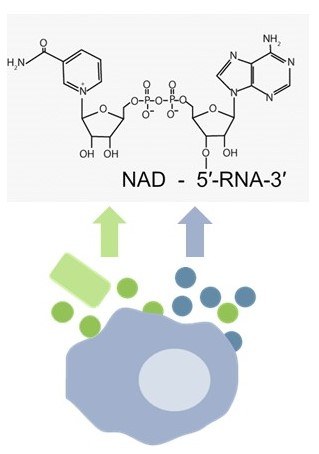
We will investigate the hypothesis that diffusible metabolites of the NAD metabolism and subsequently NAD-RNA transcripts have an important influence on the infection of macrophages with Klebsiella pneumoniae. NAD is an important coenzyme of many biological processes and regulates in particular gene expression and redox reactions. Diffusible NAD precursor molecules such as nicotinamide and nicotinic acid (vitamin B3) influence cellular NAD levels and NAD biosynthetic pathways are therapeutic targets for human diseases.
The importance of NAD in bacterial infection is illustrated by recently discovered toxins.
In this project we will analyze how the abundance of diffusible NAD precursors (nicotinamide, vitamin B3), NAD and NAD-RNA transcripts changes during the course of infection and how NAD metabolism influences the course of macrophage infection and reveals new targets for infection control.
D2 - Interspecies RNA Networks
PI: Leon Schulte
K. pneumoniae is one of the most common hospital germs and assumes new antibiotic resistance relatively quickly. As a result, hospitalized patients increasingly suffer from untreatable infections. The course of such infections is determined by mutual adaptations and reactions of host and pathogen. Using the Dual RNA-Seq method developed by us, we can reconstruct this interplay at the level of cross-species RNA networks. Our Dual RNA-Seq preliminary work with K. pneumoniae shows on the one hand that macrophages react differently after contact with the pathogen than with apathogenic bacterial stimuli, and on the other hand that the pathogen massively upregulates several transport systems for the absorption of diffusible substances such as heme, sulfates, amino acids, sugars, inositol and cobalamins. We will investigate which RNA regulatory circuits control the absorption of diffusible substances and thus the host colonization. This work will reveal urgently needed targets for the treatment of K. pneumoniae infections.
D3 - Integrated Modeling of Host-Microbe-Interactions
PI: Alexander Goesmann
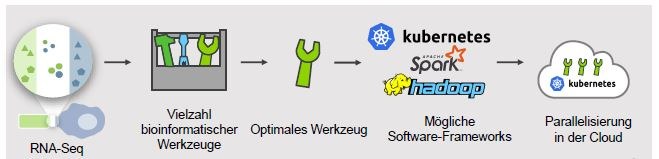
The integration of cross-species data sets from monocytes/macrophages and Gram-negative bacteria for the characterization of adaptation strategies using diffusible signals requires new analytical methods that can efficiently process large, heterogeneous data sets and model complex interaction networks between host and pathogen. This requires new approaches to deal with the ever faster growing data volumes with standardized and scalable analysis pipelines. When applying the workflows to experimental data sets, the FAIR principles must be considered in order to ensure the long-term re-use of the data.
The general goal of this subproject is therefore to support our cooperation partners within this consortium by implementing fully automated and well standardized analysis workflows that are user-friendly and efficiently scalable in cloud computing environments.




